Expression Datasets
01 Alphonso Multiple Tissues
RNA-seq of Mangifera indica plant tissues from cv. Alphonso
Experimental Conditions (1 replicate):
- Root
- Bark
- Leaf
- Flower
- Peel
- Pulp
- Seed
cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Hiseq 3000 and normalized to TPM. This dataset was published by Wang et al. 2020 and raw data can be found in the BioProject PRJNA487154.
02 Fruit Pulp and Peel Ripening
RNA-seq of Mangifera indica pulp and peel on unripe and ripe stages of the varieties Hongyu, Guire-82, and Sensation, which show different coloration during maturation stages.
Experimental Conditions (3 replicates):
- Unripe peel and pulp of cv. Hongyu (Hon) and Guire-82 (Gui)
- Ripe Peel and pulp of cv. Hongyu (Hon) and Guire-82 (Gui)
- Peel and pulp at 94, 100, 106 and 112 days after pollination (DAP) of cv. Sensation
94, 100, 106 and 112 DAP are equivalent to 2, 8, 14, 20 days not-bagged fruit.
cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Hiseq 3000 and normalized to TPM.
This dataset was published by Wang et al. 2020
and raw data can be found in the BioProject PRJNA487154.








Unripe and ripe stages of fruits of the varieties Hongyu, and Guire-82, and fruits at 94, 100, 106 and 112 days after pollination (DAP) of cv. Sensation
DAP = Days After Pollination
03 Kent Pulp Ripening
RNA-seq of Mangifera indica (cultivar Kent) pulp (mesocarp). Sampling was done at day 1 (mature-green mango) and day 10 (ripe mango), using two individual fruits for each ripening stage.
Experimental Conditions (2 replicates):
- Pulp unripe mango (mature-green mango).
- Pulp ripe mango.
Mango fruit cultivar “Kent” was harvested at the National Institute of Forestry, Agricultural and Veterinary Research (INIFAP) research station located in Navojoa, Sonora, Mexico (27°03′49.33′′ N and 109°30′11.42′′ W). Fruits were disinfected with chlorinated water and stored at 20°C up to 10 days. cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Hiseq 2000 and normalized to TPM. This dataset was published by Dautt-Castro et al. 2015 and raw data can be found in the BioProject PRJNA258477 .
04 Keitt Peel Ripening
RNA-seq of Mangifera indica (cultivar Keitt) peel in ripe and overripe stages to study cuticle biosynthesis.
Experimental Conditions (3 replicates):
- Peel ripe.
- Peel overripe.
Mango (Mangifera indica, cultivar Keitt) fruit used for RNA-Seq profiling were obtained from a commercial store at Ithaca, New York, USA. The fruit were divided into two stages: ripe and overripe and were stored at room temperature (approximately 20 °C and 60–65% relative humidity) for 12 days. After ripening, the fruit were stored at 20 °C and 60–65% relative humidity for 18 days, with sampling carried out every six days. Peel samples from three fruit were pooled to create a single biological replicate and three independent biological replicates of both ripe and overripe fruit were analyzed. cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Hiseq 2500 and normalized to TPM. This dataset was published by Tafolla-Arellano et al. 2017 and raw data can be found in the BioProject PRJNA253272.


Mango fruit at the stages used for RNA-Seq analysis: ripe (left) and overripe (right).
05 Keitt Peel Storage
RNA-seq of Mangifera indica (cultivar Keitt) peel at harvest and after 2, 7, and 14 days of cold storage at 5 or 12°C, to study storage chilling stress.
Experimental Conditions (2 replicates):
- 0 days harvest.
- 2,7,14 days after harvest (DAH) stored at 5 celsius.
- 2,7,14 days after harvest (DAH) stored at 12 celsius.
Mango (Mangifera indica, cultivar Keitt) were obtained from a commercial storage house (Mor Hasharon, Israel) 1–2 h after harvest and transported (1 h) to the Agricultural Research Organization (Israel). Uniform, unblemished fruit weighing 424 ± 16 g were selected. To remove the fruit sap, the fruit was dipped into water after harvest; no other treatment was applied after harvest. Peel tissue (±5 g) was randomly sliced from six fruit per biological replicate at harvest and after 2, 7, and 14 days of cold storage at 5 or 12°C, each with two biological replicates. cDNA libraries were sequenced by SE sequencing with Illumina Hiseq 2000 and normalized to TPM. This dataset was published by Sivankalyani et al. 2016 and raw data can be found in the BioProject PRJNA304093.




Mango fruits showing chilling injury symptoms after 19 days of cold storage at 5°C (left) and 12°C (right).
DAH = Days After Harvest
06 Tainong and Renong Pulp Ripening
RNA-seq of Mangifera indica pulp from two mango cultivars with different sweetness (high-sweet mango Tainong-1 and low-sweet mango Renong-1) were used to study fruit ripening.
Experimental Conditions (3 replicates):
- Renong pulp 30 days after pollination (dap) - Renong-1 unripe
- Renong pulp 95 dap - Renong-1 early ripe
- Renong pulp 130 dap - Renong-1 partially ripe
- Renong pulp fully ripe
- Tainong pulp 30 dap - Tainong-1 unripe
- Tainong pulp 60 dap - Tainong-1 early ripe
- Tainong pulp 100 dap - Tainong-1 partially ripe
- Tainong pulp fully ripe
Fruit samples were collected at four ripening stages including unripe (~ 30 days after pollination), early ripe (~ 60 days after pollination), partial ripe (~ 100 days after pollination) and ripe (~ 110 days after pollination) from 3 years old plants held at the experimental station of the South Subtropical Crops Research Institute of the Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China. Plants were grown in natural environment and five mixed fruit samples per plant were collected from three different plants of each cultivar (three biological replicates). cDNA libraries were sequenced by PE sequencing (300 x 2) with Illumina HiSeq 2500 and normalized to TPM. This dataset was published by Li et al. 2020 and raw data can be found in the BioProject PRJNA629065.








Changes in mango fruit during ripening. From left to right 4 pictures of mango cultivar Renong-1 fruits (top), and Tainong-1 (bottom) at unripe, early ripe, partially ripe and ripe stages.
DAP = Days After Pollination
07 Tainong Pulp Ripening
RNA-seq of Mangifera indica pulp (mesocarp) fruit ripening from cv. Tainong
Experimental Conditions (3 replicates):
- Tainong pulp of fruits harvested at 40, 60, 80, and 90 days after flowering (DAF)
- Tainong pulp of fruits harvested at 90 days and stored for 0, 4, 8 and 12 days after harvest (DAH)
Samples were collected from the nursery at different stages of fruit development (40, 60, 80, and 90 days after the flowering stage began). Fruit samples harvested at 90 days after the flowering were also kept for 4, 8, and 12 days in the laboratory at a controlled temperature of 25°C and a humidity level of 95%. Seven mangoes of an average weight of 100 ± 10 g were sampled. The fruits were picked from different trees at the same positions on the day of fruit collection. cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina HiSeq 2500 and normalized to TPM. This dataset was published by Xin et al. 2021 and raw data can be found in the BioProject PRJNA697524.






From left to right, morphology of mango fruits at 40, 60, 80 days after flowering (DAF), and 4, 8 and 12 days stored after harvest (DAH).
Samples labeled 40 DAF, 60 DAF, and 80 DAF are pulp samples of fruits harvested at 40, 60, and 80 days after flowering, respectively.
The sample labeled 0 DAH, corresponds to pulp of fruits harvested at 90 days after flowering.
Samples labeled 4 DAH, 8 DAH, and 12 DAH are pulp samples of fruits harvested at 90 days after flowering and stored for 4, 8, and 12 days, respectively.
DAF = Days After Flowering
DAH = Days After harvest (harvested after 90 DAF)
08 Chokanan Pulp Ripening
RNA-seq of Mangifera indica pulp (mesocarp) fruit ripening from cv. Chokanan, at immature and ripening stages within the same season.
Experimental Conditions (2 replicates):
- Unripe pulp 21 Days After Pollination (DAP)
- Ripe pulp 75 DAP
Fruits were harvested from two trees that were grown on the same soil and from the same rootstock, thus minimizing any external factor and genetic variation that may affects the fruit development. The fruits were harvested at 21 Days After Pollination (DAP) for immature stage, and at 75 DAP for the ripening stage. The fruits were dissected by removing the skin where the mesocarp was cut into 1 cm cubes. Two biological replicate of mango fruits mesocarp were sequences from each stages. cDNA libraries were sequenced by SE sequencing with Illumina Hiseq 4000 and normalized to TPM. This dataset was published by Karim. et al. 2022 and raw data can be found in the BioProject PRJNA803945.
DAP = Days After Pollination
09 Chokanan and Golden Phoenix Pulp Ripening
RNA-seq of Mangifera indica pulp from varieties Chokanan (firm), and Golden phoenix and Water lily (less-firm pulp).
Experimental Conditions (3 replicates):
- Chokanan ripe mango.
- Chokanan unripe mango.
- Golden phoenix + Water lily ripe mango.
- Golden phoenix + Water lily unripe mango.
Mango fruit varieties (‘Chokanan’, ‘Golden phoenix’ and ‘Water lily’) were obtained from a commercial supplier at the mature green stage. Following this, fruits were washed in running tap water, air dried and allowed to ripen at ambient temperature (25 ± 1 °C). Fruits were sampled at the unripe and ripe stages, using three individual fruits per ripening stage. Equal amounts of RNA samples from the less-firm mango varieties ‘Golden phoenix’ and ‘Water lily’ were combined to represent the less-firm mango group. cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Hiseq 2000 and normalized to TPM. This dataset was published by Lawson et al. 2020 and raw data can be found in the BioProject PRJNA515564.
10 Shelly Peel Hot Water Treatment
RNA-seq of Mangifera indica (cultivar Shelly) peel after 0, 4, 17 and 48 hours following hot water brush (HWB) post-harvest treatment and their controls (no post-harvest treatment).
Experimental Conditions (These experiments only contain 1 replicate):
- Peel 0, 4, 17 and 48 hours controls.
- Peel 0, 4, 17 and 48 hours following postharvest hot water treatment.
Postharvest treatment by hot water brushing (HWB) for 15–20 s was introduced commercially to improve fruit quality and reduce postharvest disease. This treatment enabled successful storage for 3–4 weeks at 12°C, with improved color and reduced disease development, but it enhanced lenticel discoloration on the fruit peel. Freshly harvested mango fruits (Mangifera indica L. cv. Shelly) were obtained from trees in commercial orchards in the north of Israel. cDNA libraries were sequenced by SE sequencing with Illumina Hiseq 2000 and normalized to TPM. This dataset was published by Luria et al. 2014 and raw data can be found in the BioProject PRJNA227243.


Example of control mango (on the left) and a mango after hot water brush post-harvest treatment (on the right).
HWB = Hot Water Brush
11 Ataulfo Pulp Quarantine Postharvest Treatment
RNA-seq of Mangifera indica (cultivar Ataulfo) pulp after 1 (unripe) and 10 (ripe) days following quarantine postharvest treatment and their controls.
Experimental Conditions (2 replicates):
- Pulp 1 and 10 days controls
- Pulp 1 and 10 days following quarantine postharvest treatment.
Mature-green mango fruits cv. Ataulfo were obtained at the Diazteca packing facility in Escuinapa, Sinaloa, México, where they were subjected to the United States Agricultural Department APHIS hot water treatment (HWT). cDNA libraries were sequenced by PE sequencing (100 x 2) with Illumina Genome Analyzer IIx and normalized to TPM. This dataset was published by Dautt-Castro et al. 2018. Raw data can be found in the BioProject PRJNA286253.
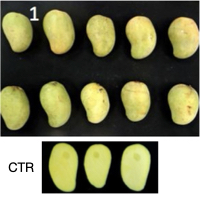


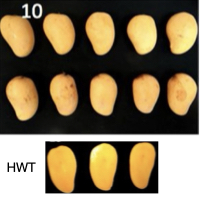
From left to right, mango fruits and pulp (below) in control conditions after 1 day post-harvest (unripe), fruits and pulp after hot water treatment (HWT) and 1 day post-harvest, fruits and pulp in control conditions and 10 days post-harvest (ripe), fruits and pulp after HWT and 10 days post-harvest.
Ata = Ataulfo
CTR = Control
HWT = Hot water treatment
12 Shelly peel Colletotrichum gloeosporioides treatment
RNA-seq of Mangifera indica (cultivar Shelly) from the peel of the red and green sides of the red and green fruit inoculated with C. gloeosporioides at time 0, 2, 7 days post inoculation.
Experimental Conditions (3 replicates):
- Peel from Green Fruit Green Side at 0, 2, 7 days post inoculation (dpi).
- Peel from Green Fruit Red Side at 0, 2, 7 dpi.
- Peel from Red Fruit Green Side at 0, 2, 7 dpi.
- Peel from Red Fruit Red Side at 0, 2, 7 dpi.
Mango fruits were harvested in July 2017. The fruit were picked from two different positions of the canopy in the orchard: the ‘red’ colored fruit (RF) from the exterior position with direct exposure to sunlight, and the ‘green’ colored fruit (GF) from the interior area in the shaded part of the canopy. cDNA libraries were sequenced by SE sequencing with Illumina HiSeq 2500 and normalized to TPM. This dataset was published by Sudheeran PK et al. 2021. Raw data can be found in the BioProject PRJNA575336 .

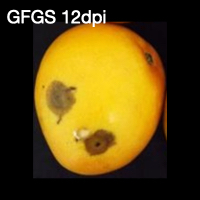
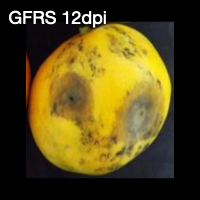



From left to right, green mango fruit 0 days post inoculation (dpi), green fruit green side (GFGS) 12 dpi, green fruit red side (GFRS) 12 dpi, red mango fruit 0 dpi, red fruit green side (RFGS) 12 dpi, and red fruit red side (RFRS) 12 dpi.
GFGS = Green Fruit Green Side
GFRS = Green Fruit Red Side
RFGS = Red Fruit Green Side
RFRS = Red Fruit Red Side
dpi = days post inoculation

